W3cubDocs
/scikit-learnsklearn.cluster.MiniBatchKMeans
-
class sklearn.cluster.MiniBatchKMeans(n_clusters=8, init='k-means++', max_iter=100, batch_size=100, verbose=0, compute_labels=True, random_state=None, tol=0.0, max_no_improvement=10, init_size=None, n_init=3, reassignment_ratio=0.01)[source] -
Mini-Batch K-Means clustering
Read more in the User Guide.
Parameters: n_clusters : int, optional, default: 8
The number of clusters to form as well as the number of centroids to generate.
max_iter : int, optional
Maximum number of iterations over the complete dataset before stopping independently of any early stopping criterion heuristics.
max_no_improvement : int, default: 10
Control early stopping based on the consecutive number of mini batches that does not yield an improvement on the smoothed inertia.
To disable convergence detection based on inertia, set max_no_improvement to None.
tol : float, default: 0.0
Control early stopping based on the relative center changes as measured by a smoothed, variance-normalized of the mean center squared position changes. This early stopping heuristics is closer to the one used for the batch variant of the algorithms but induces a slight computational and memory overhead over the inertia heuristic.
To disable convergence detection based on normalized center change, set tol to 0.0 (default).
batch_size : int, optional, default: 100
Size of the mini batches.
init_size : int, optional, default: 3 * batch_size
Number of samples to randomly sample for speeding up the initialization (sometimes at the expense of accuracy): the only algorithm is initialized by running a batch KMeans on a random subset of the data. This needs to be larger than n_clusters.
init : {‘k-means++’, ‘random’ or an ndarray}, default: ‘k-means++’
Method for initialization, defaults to ‘k-means++’:
‘k-means++’ : selects initial cluster centers for k-mean clustering in a smart way to speed up convergence. See section Notes in k_init for more details.
‘random’: choose k observations (rows) at random from data for the initial centroids.
If an ndarray is passed, it should be of shape (n_clusters, n_features) and gives the initial centers.
n_init : int, default=3
Number of random initializations that are tried. In contrast to KMeans, the algorithm is only run once, using the best of the
n_initinitializations as measured by inertia.compute_labels : boolean, default=True
Compute label assignment and inertia for the complete dataset once the minibatch optimization has converged in fit.
random_state : integer or numpy.RandomState, optional
The generator used to initialize the centers. If an integer is given, it fixes the seed. Defaults to the global numpy random number generator.
reassignment_ratio : float, default: 0.01
Control the fraction of the maximum number of counts for a center to be reassigned. A higher value means that low count centers are more easily reassigned, which means that the model will take longer to converge, but should converge in a better clustering.
verbose : boolean, optional
Verbosity mode.
Attributes: cluster_centers_ : array, [n_clusters, n_features]
Coordinates of cluster centers
labels_ : :
Labels of each point (if compute_labels is set to True).
inertia_ : float
The value of the inertia criterion associated with the chosen partition (if compute_labels is set to True). The inertia is defined as the sum of square distances of samples to their nearest neighbor.
See also
-
KMeans - The classic implementation of the clustering method based on the Lloyd’s algorithm. It consumes the whole set of input data at each iteration.
Notes
See http://www.eecs.tufts.edu/~dsculley/papers/fastkmeans.pdf
Methods
fit(X[, y])Compute the centroids on X by chunking it into mini-batches. fit_predict(X[, y])Compute cluster centers and predict cluster index for each sample. fit_transform(X[, y])Compute clustering and transform X to cluster-distance space. get_params([deep])Get parameters for this estimator. partial_fit(X[, y])Update k means estimate on a single mini-batch X. predict(X)Predict the closest cluster each sample in X belongs to. score(X[, y])Opposite of the value of X on the K-means objective. set_params(**params)Set the parameters of this estimator. transform(X[, y])Transform X to a cluster-distance space. -
__init__(n_clusters=8, init='k-means++', max_iter=100, batch_size=100, verbose=0, compute_labels=True, random_state=None, tol=0.0, max_no_improvement=10, init_size=None, n_init=3, reassignment_ratio=0.01)[source]
-
fit(X, y=None)[source] -
Compute the centroids on X by chunking it into mini-batches.
Parameters: X : array-like, shape = [n_samples, n_features]
Coordinates of the data points to cluster
-
fit_predict(X, y=None)[source] -
Compute cluster centers and predict cluster index for each sample.
Convenience method; equivalent to calling fit(X) followed by predict(X).
-
fit_transform(X, y=None)[source] -
Compute clustering and transform X to cluster-distance space.
Equivalent to fit(X).transform(X), but more efficiently implemented.
-
get_params(deep=True)[source] -
Get parameters for this estimator.
Parameters: deep: boolean, optional :
If True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns: params : mapping of string to any
Parameter names mapped to their values.
-
partial_fit(X, y=None)[source] -
Update k means estimate on a single mini-batch X.
Parameters: X : array-like, shape = [n_samples, n_features]
Coordinates of the data points to cluster.
-
predict(X)[source] -
Predict the closest cluster each sample in X belongs to.
In the vector quantization literature,
cluster_centers_is called the code book and each value returned bypredictis the index of the closest code in the code book.Parameters: X : {array-like, sparse matrix}, shape = [n_samples, n_features]
New data to predict.
Returns: labels : array, shape [n_samples,]
Index of the cluster each sample belongs to.
-
score(X, y=None)[source] -
Opposite of the value of X on the K-means objective.
Parameters: X : {array-like, sparse matrix}, shape = [n_samples, n_features]
New data.
Returns: score : float
Opposite of the value of X on the K-means objective.
-
set_params(**params)[source] -
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.Returns: self :
-
transform(X, y=None)[source] -
Transform X to a cluster-distance space.
In the new space, each dimension is the distance to the cluster centers. Note that even if X is sparse, the array returned by
transformwill typically be dense.Parameters: X : {array-like, sparse matrix}, shape = [n_samples, n_features]
New data to transform.
Returns: X_new : array, shape [n_samples, k]
X transformed in the new space.
-
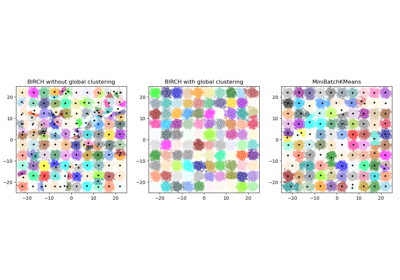
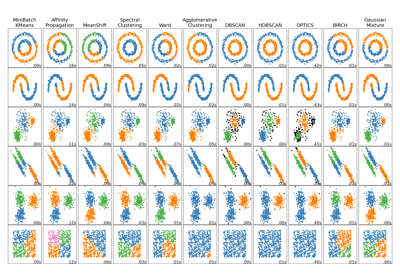
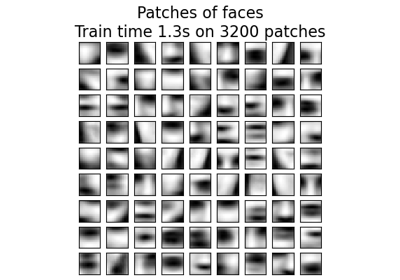
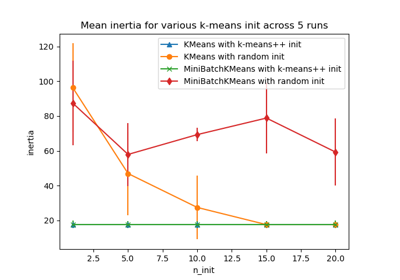
Examples using sklearn.cluster.MiniBatchKMeans
© 2007–2016 The scikit-learn developers
Licensed under the 3-clause BSD License.
http://scikit-learn.org/stable/modules/generated/sklearn.cluster.MiniBatchKMeans.html