W3cubDocs
/scikit-learnConcentration Prior Type Analysis of Variation Bayesian Gaussian Mixture
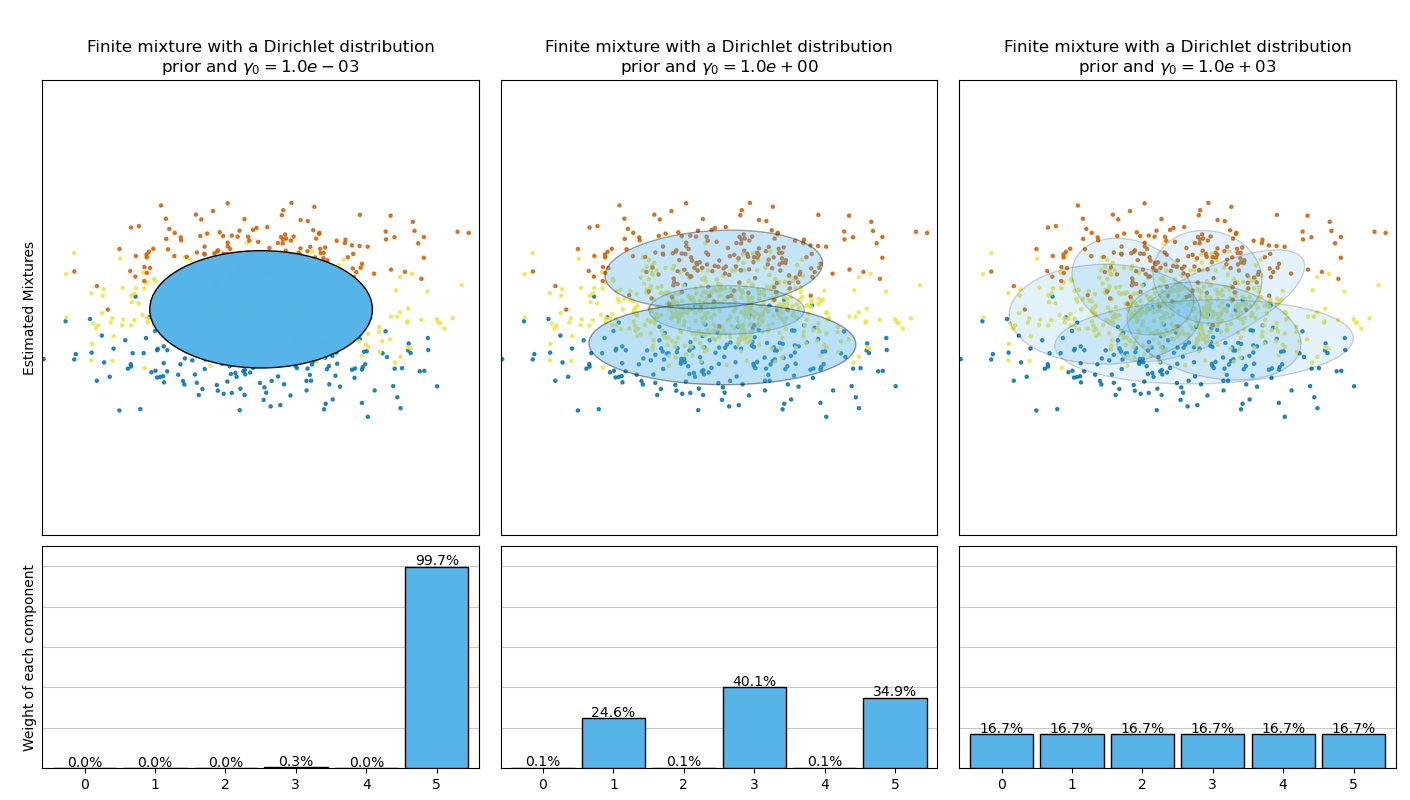
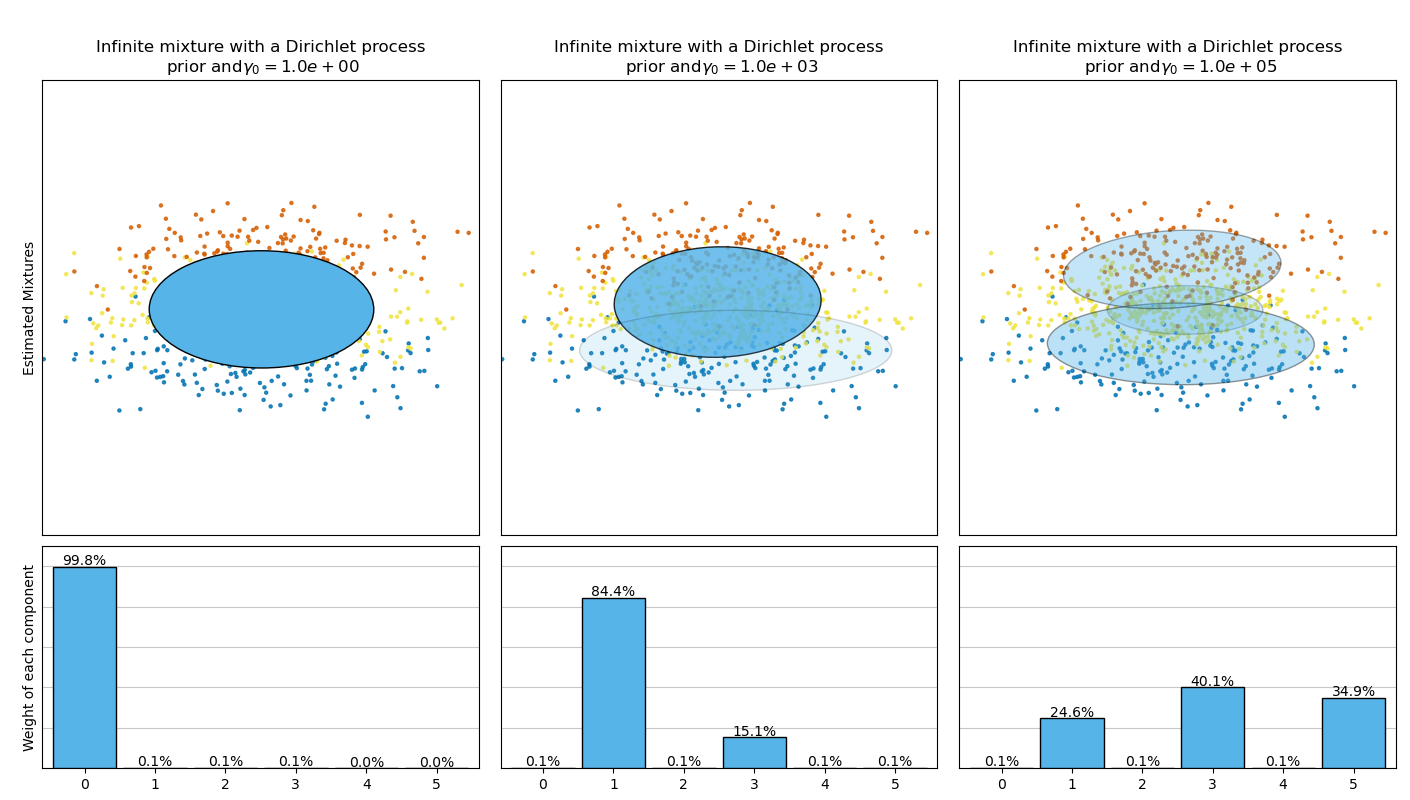
This example plots the ellipsoids obtained from a toy dataset (mixture of three Gaussians) fitted by the BayesianGaussianMixture class models with a Dirichlet distribution prior (weight_concentration_prior_type='dirichlet_distribution') and a Dirichlet process prior (weight_concentration_prior_type='dirichlet_process'). On each figure, we plot the results for three different values of the weight concentration prior.
The BayesianGaussianMixture class can adapt its number of mixture componentsautomatically. The parameter weight_concentration_prior has a direct link with the resulting number of components with non-zero weights. Specifying a low value for the concentration prior will make the model put most of the weight on few components set the remaining components weights very close to zero. High values of the concentration prior will allow a larger number of components to be active in the mixture.
The Dirichlet process prior allows to define an infinite number of components and automatically selects the correct number of components: it activates a component only if it is necessary.
On the contrary the classical finite mixture model with a Dirichlet distribution prior will favor more uniformly weighted components and therefore tends to divide natural clusters into unnecessary sub-components.
# Author: Thierry Guillemot <[email protected]> # License: BSD 3 clause import numpy as np import matplotlib as mpl import matplotlib.pyplot as plt import matplotlib.gridspec as gridspec from sklearn.mixture import BayesianGaussianMixture print(__doc__) def plot_ellipses(ax, weights, means, covars): for n in range(means.shape[0]): eig_vals, eig_vecs = np.linalg.eigh(covars[n]) unit_eig_vec = eig_vecs[0] / np.linalg.norm(eig_vecs[0]) angle = np.arctan2(unit_eig_vec[1], unit_eig_vec[0]) # Ellipse needs degrees angle = 180 * angle / np.pi # eigenvector normalization eig_vals = 2 * np.sqrt(2) * np.sqrt(eig_vals) ell = mpl.patches.Ellipse(means[n], eig_vals[0], eig_vals[1], 180 + angle) ell.set_clip_box(ax.bbox) ell.set_alpha(weights[n]) ell.set_facecolor('#56B4E9') ax.add_artist(ell) def plot_results(ax1, ax2, estimator, X, y, title, plot_title=False): ax1.set_title(title) ax1.scatter(X[:, 0], X[:, 1], s=5, marker='o', color=colors[y], alpha=0.8) ax1.set_xlim(-2., 2.) ax1.set_ylim(-3., 3.) ax1.set_xticks(()) ax1.set_yticks(()) plot_ellipses(ax1, estimator.weights_, estimator.means_, estimator.covariances_) ax2.get_xaxis().set_tick_params(direction='out') ax2.yaxis.grid(True, alpha=0.7) for k, w in enumerate(estimator.weights_): ax2.bar(k - .45, w, width=0.9, color='#56B4E9', zorder=3) ax2.text(k, w + 0.007, "%.1f%%" % (w * 100.), horizontalalignment='center') ax2.set_xlim(-.6, 2 * n_components - .4) ax2.set_ylim(0., 1.1) ax2.tick_params(axis='y', which='both', left='off', right='off', labelleft='off') ax2.tick_params(axis='x', which='both', top='off') if plot_title: ax1.set_ylabel('Estimated Mixtures') ax2.set_ylabel('Weight of each component') # Parameters of the dataset random_state, n_components, n_features = 2, 3, 2 colors = np.array(['#0072B2', '#F0E442', '#D55E00']) covars = np.array([[[.7, .0], [.0, .1]], [[.5, .0], [.0, .1]], [[.5, .0], [.0, .1]]]) samples = np.array([200, 500, 200]) means = np.array([[.0, -.70], [.0, .0], [.0, .70]]) # mean_precision_prior= 0.8 to minimize the influence of the prior estimators = [ ("Finite mixture with a Dirichlet distribution\nprior and " r"$\gamma_0=$", BayesianGaussianMixture( weight_concentration_prior_type="dirichlet_distribution", n_components=2 * n_components, reg_covar=0, init_params='random', max_iter=1500, mean_precision_prior=.8, random_state=random_state), [0.001, 1, 1000]), ("Infinite mixture with a Dirichlet process\n prior and" r"$\gamma_0=$", BayesianGaussianMixture( weight_concentration_prior_type="dirichlet_process", n_components=2 * n_components, reg_covar=0, init_params='random', max_iter=1500, mean_precision_prior=.8, random_state=random_state), [1, 1000, 100000])] # Generate data rng = np.random.RandomState(random_state) X = np.vstack([ rng.multivariate_normal(means[j], covars[j], samples[j]) for j in range(n_components)]) y = np.concatenate([j * np.ones(samples[j], dtype=int) for j in range(n_components)]) # Plot results in two different figures for (title, estimator, concentrations_prior) in estimators: plt.figure(figsize=(4.7 * 3, 8)) plt.subplots_adjust(bottom=.04, top=0.90, hspace=.05, wspace=.05, left=.03, right=.99) gs = gridspec.GridSpec(3, len(concentrations_prior)) for k, concentration in enumerate(concentrations_prior): estimator.weight_concentration_prior = concentration estimator.fit(X) plot_results(plt.subplot(gs[0:2, k]), plt.subplot(gs[2, k]), estimator, X, y, r"%s$%.1e$" % (title, concentration), plot_title=k == 0) plt.show()
Total running time of the script: (0 minutes 17.509 seconds)
plot_concentration_prior.py
plot_concentration_prior.ipynb
© 2007–2016 The scikit-learn developers
Licensed under the 3-clause BSD License.
http://scikit-learn.org/stable/auto_examples/mixture/plot_concentration_prior.html